Membrane proteins are encoded by around 20% of the human genome and constitute a majority of drug targets, but they only account for about 3% of known protein structures. NMR spectroscopy can contribute to our understanding of membrane protein structure and function, since it is able to study functional dynamics and conformational transitions in membrane proteins embedded in native-like lipid bilayer environments, such as liposomes and lipid nanodiscs. We study both transmembrane and extramembrane regions of membrane proteins using solid- and solution-state NMR methodology.
In a collaborative project headed by Dr. Françoise Jacob-Dubuisson (Lille Center for Infection and Immunity (CIIL), Institut Pasteur Lille), we study the 61 kDa bacterial outer membrane protein FhaC. This 16-stranded beta-barrel protein serves to transport the virulence factor FHA across the outer membrane of the bacterium Bordetella pertussis, the causative agent of whooping cough. Its resting-state three-dimensional structure is known [1], but alternative conformations assumed during the transport cycle, as well as its general transport mechanism, have so far remained elusive. We investigate wild type FhaC, as well as mutant variants affecting its transport properties, reconstituted into liposomes using solid-state NMR as well as in nanodiscs via solution-state NMR. Specific labeling of isoleucine methyls is used to reduce spectral overlap in this large protein. We apply exchange NMR methods such as R1rho relaxation dispersion, as well as paramagnetic relaxation enhancements, to identify alternative FhaC conformations and their exchange dynamics.
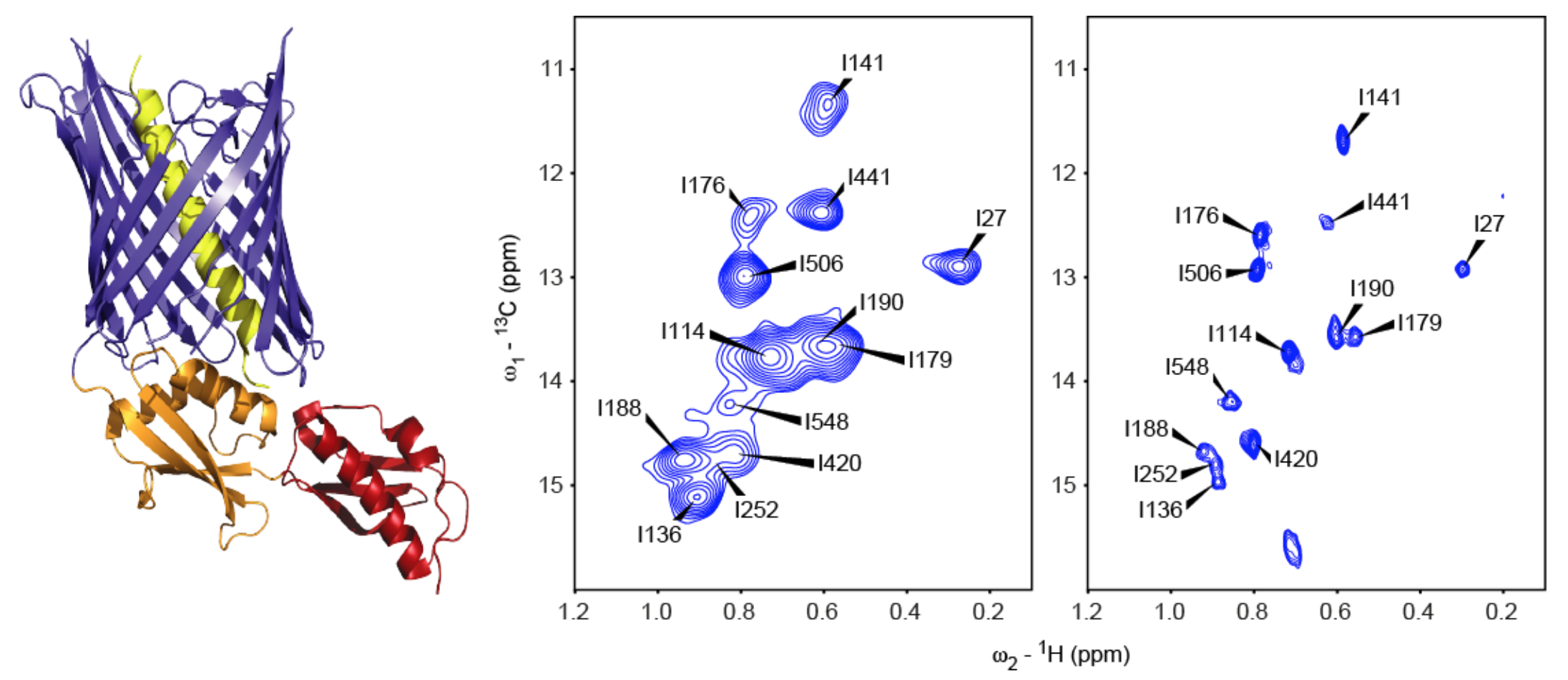
Fig. 1 - Left: Resting-state crystal structure of FhaC (PDB 4QKY, [1]). Blue: transmembrane beta-barrel; red/orange: periplasmic POTRA (polypeptide transport associated) domains; yellow: N-terminal alpha-helix. Center: Solid-state dipolar hCH correlation spectrum of isoleucine δ1-methyl (13C,1H)-labeled FhaC in liposomes. Right: Solution-state SOFAST-HMQC spectrum of isoleucine δ1-methyl (13C,1H)-labeled FhaC in nanodiscs.
Together with the group of Prof. Petri Kursula at the University of Bergen (Norway), we study the P0 membrane protein of the myelin sheath of the peripheral nervous system. P0 is involved in the tight stacking of membranes characteristic of myelin, both via its extracellular Ig-like domain and its cytoplasmic disordered C-terminal tail (P0ct), and mutations in P0 are implicated in demyelinating diseases such as Charcot-Marie-Tooth disease [2]. We specifically investigate the disordered P0ct region, its conformational sampling in solution as well as its folding-upon-binding during interaction with lipid bilayers, both with solution- and solid-state NMR spectroscopy.
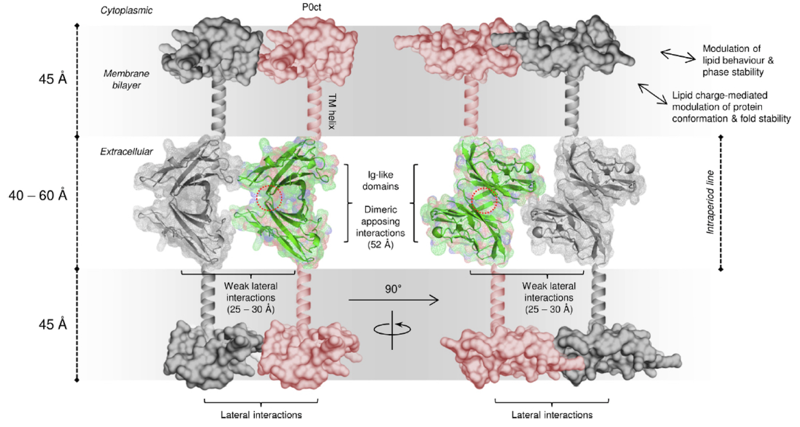
Fig. 2 - Model for the ultrastructure of the myelin intraperiod line with myelin protein P0. Apposing extracellular P0 Ig-like domains interact across the extracellular space (center), while the disordered C-terminal domain anchors onto the cytoplasmic phospholipid headgroups (top and bottom), potentially regulating lateral P0 spacing and affecting bilayer properties. Figure taken from Raasakka et al. [2].
Voltage-gated ion channels (VGICs) confer electrical excitability to cells and are pivotal in processes such as neuronal communication, heartbeat, and motility. Much structural and functional work has provided a wealth of information on these membrane proteins, notably numerous three-dimensional structures. However, the resting state of their voltage-sensitive domains (VSDs) that translate the membrane potential into structural changes has so far largely remained elusive, since it is only assumed in the presence of the cell's resting membrane potential, which is absent from samples used in crystallographic or cryo-electron microscopic structure determination [3]. Solid-state NMR spectroscopy can potentially circumvent this problem, since it can study membrane proteins in liposomes, which provide both a lipid bilayer environment and a closed compartment. We attempt to generate a stable resting transmembrane potential in liposomes via the asymmetric presence of large impermeable polyions, in order to stabilize the resting state of the VSDs of VGICs reconstituted into such liposomes and to then study its structure using solid-state NMR.
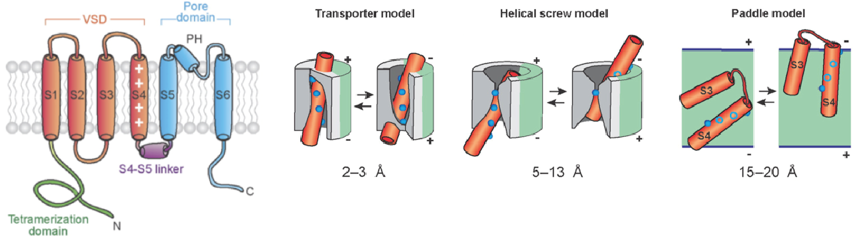
Fig. 3 - Left: Architecture of a VGIC channel subunit. Cylinders are helical segments. Blue: pore domain; red: voltage-sensing domain (VSD); purple: linker between S4-S5 helices; green: N-terminal tetramerization domain. Right: Illustrations of different models of voltage sensing by VSDs. Red cylinders: VSD helices; blue spheres: arginine residues; gray: surrounding protein; green: lipid. In each model, the resting position of the S4 helix is shown on the left, and the activated position on the right. The extent of S4 transmembrane motion involved is reported below each model. Figure adapted from Tombola et al. [3].
Funding:
Agence Nationale de Recherche, Projet de Recherche Collaborative grant "OPENBAR: Functional conformations of the protein transporter FhaC" (ANR-17-CE11-0043-02)
Agence Nationale de Recherche, Jeunes Chercheuses Jeunes Chercheurs grant "CREST: The resting state of voltage-gated ion channels by NMR" (ANR-19-CE11-0006-01)
References
[1] Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 2007, 317, 957-961. DOI: 10.1126/science.1143860
[2] Raasakka A, Ruskamo S, Kowal J, Han H, Baumann A, Myllykoski M, Fasano A, Rossano R, Riccio P, Bürck J, Ulrich AS, Stahlberg H, Kursula P. Molecular structure and function of myelin protein P0 in membrane stacking. Sci. Rep. 2019, 9, 642. DOI: 10.1038/s41598-018-37009-4
[3] Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu. Rev. Cell Dev. Biol 2006, 22, 23-52. DOI: 10.1146/annurev.cellbio.21.020404.145837
Contact: robert.schneider@univ-lille.fr
